The journey of spatial transcriptomics
Unlocking the mysteries of tumor tissue architecture
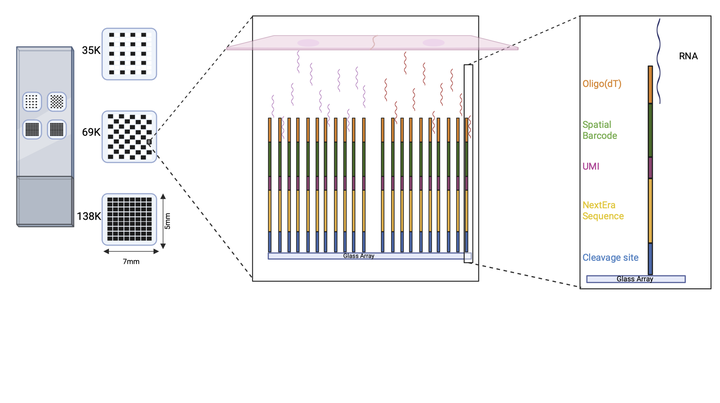 Our platform consists of a slide with four subarrays containing thousands of spots, each with a few million barcoded oligonucleotides with the following structure: anchored oligodT, spatial barcode, UMI, sequencing handle and a cleavage site. The tissue section is mounted on the array and permeabilized. Next, the released polyadenylated transcripts are captured by the oligodT domain and in situ reverse transcription is performed. After probe cleavage, second strand cDNA is synthesized and amplified, followed by cleanup and massively parallel sequencing. Finally, each sequencing read can be mapped back to its original position in the array based on the spatial barcode.
Our platform consists of a slide with four subarrays containing thousands of spots, each with a few million barcoded oligonucleotides with the following structure: anchored oligodT, spatial barcode, UMI, sequencing handle and a cleavage site. The tissue section is mounted on the array and permeabilized. Next, the released polyadenylated transcripts are captured by the oligodT domain and in situ reverse transcription is performed. After probe cleavage, second strand cDNA is synthesized and amplified, followed by cleanup and massively parallel sequencing. Finally, each sequencing read can be mapped back to its original position in the array based on the spatial barcode.
Our lab is at the forefront of innovation, tirelessly working on the development of a groundbreaking platform for spatial transcriptomics studies. This novel platform holds the promise of improving our understanding of tissue organization and function, offering enhanced resolution and capture efficiency at a more accessible price point. However, as with any pioneering endeavor, the path to making this technology available is riddled with challenges. Today, we're excited to share with you an overview of the development of this technology, its current status, and our vision for its future.
But first, let’s delve into what exactly spatial transcriptomics entails.
Understanding spatial transcriptomics
Genes, as we know, operate not just in isolation but within the intricate framework of spatial organization within tissues. From embryonic development to the complexities of tumor microenvironments, the spatial arrangement of cells plays a crucial role in determining functionality and behavior. Spatial transcriptomics seeks to illuminate this intricate relationship by providing a means to quantify RNA molecules within tissue sections while preserving their spatial context.
The emergence of spatial transcriptomics
Early attempts to visualize gene expression in a spatial context relied on techniques such as in situ hybridization (ISH), with whole-mount ISH emerging as a favored method in the '90s and early '20s. However, limitations such as the need for stereotypical tissue structures and qualitative results hindered its widespread adoption. Subsequent advances in single-cell genomic technologies offered deeper insights into cellular diversity but at the cost of losing spatial information.
Spatial transcriptomics bridges this gap by enabling transcriptome-wide analysis within intact tissue sections. This approach allows researchers to tackle a myriad of biological questions, from deciphering cell-type compositions to unraveling intricate cellular interactions and molecular dynamics within tissues.
At present, leading commercial tools such as Visium by 10X Genomics and Merfish by Vizgen dominate the spatial transcriptomics landscape. While each employs distinct methodologies, both offer valuable insights into tissue organization and function. However, there's still ample room for improvement in terms of resolution, detection efficiency, and cost-effectiveness. Furthermore, these commercial platforms are closed-box systems, making it difficult or impossible to optimize library preparation protocols or to profile other -omics layers (e.g. DNA mutation or methylation).
Introducing our innovative platform
In our quest to push the boundaries of spatial transcriptomics, we're proud to introduce our cutting-edge platform. In its current version, it is designed to capture polyadenylated transcripts from tissue sections on in situ arrays. Our platform has several key advantages:
- Enhanced resolution: With array designs featuring up to 138K squared spots of 10x10 µm (with pitch distance of 11 µm) and a capture area of 7x5 mm, we obtain significantly higher resolution compared to many existing technologies.
- Cost-effectiveness: our custom-printed slides are a fraction of the cost of current options, making spatial transcriptomics more accessible to researchers worldwide.
- Innovative synthesis method: Leveraging a light-directed synthesis method, our collaborator Prof. Mark Somoza has optimized the technology for higher throughput and cost efficiency, albeit with higher error rates (see figure below for details on the synthesis).
- Decoding precision: Through pioneering decoding algorithms developed in collaboration with Prof. William Press from the University of Texas, we ensure high precision (and acceptable recall) in spatial -omics data analysis.
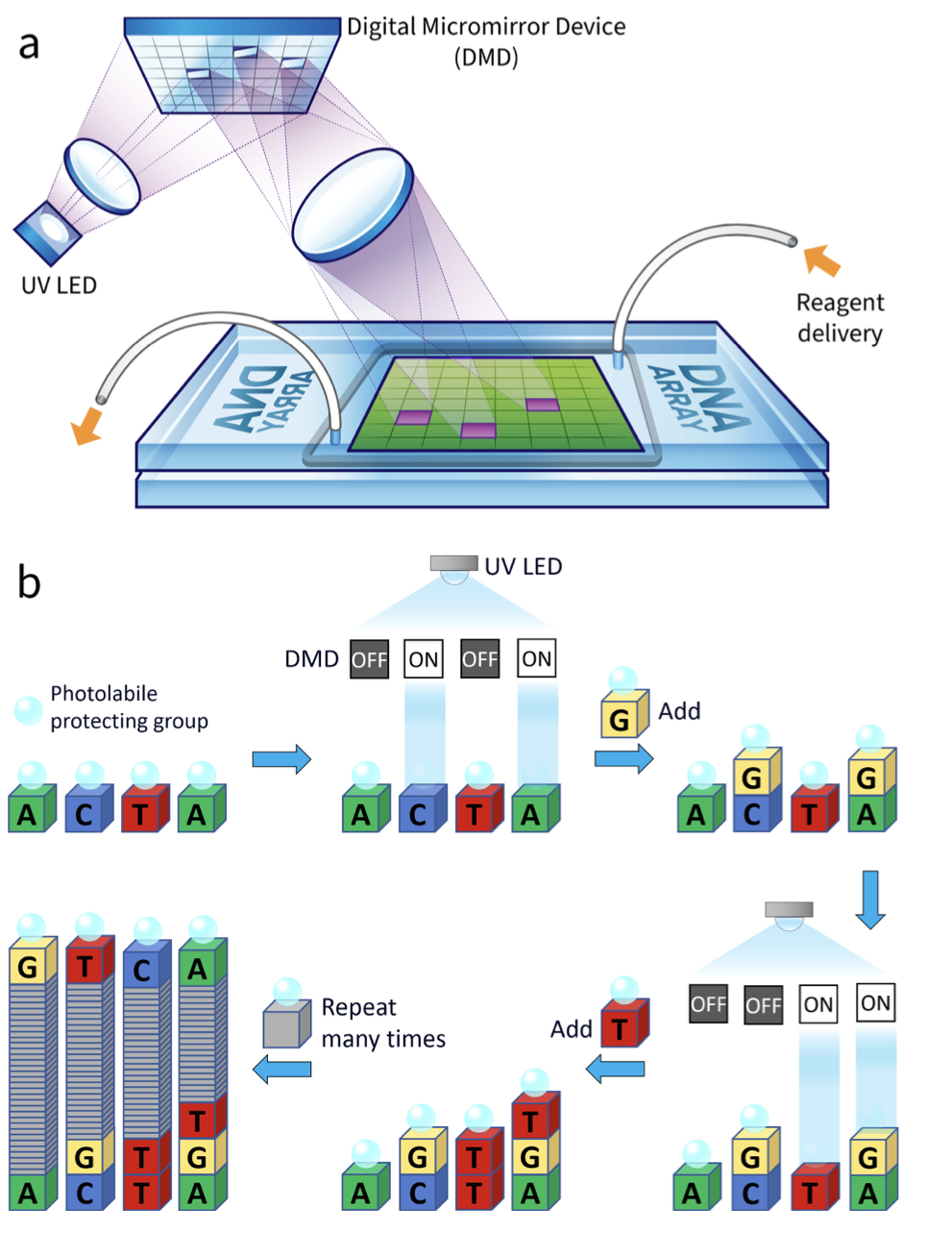 Ultraviolet light is imaged onto the digital micromirror device (DMD), and reflections from the ON mirrors are further imaged onto a glass substrate mounted in a flow cell connected to a standard solid phase oligonucleotide synthesizer, as shown schematically in the figure. Each light exposure is then followed by a coupling reaction that results in a sequence‐defined synthesis of oligonucleotides, with one sequence per mirror
(source)
Ultraviolet light is imaged onto the digital micromirror device (DMD), and reflections from the ON mirrors are further imaged onto a glass substrate mounted in a flow cell connected to a standard solid phase oligonucleotide synthesizer, as shown schematically in the figure. Each light exposure is then followed by a coupling reaction that results in a sequence‐defined synthesis of oligonucleotides, with one sequence per mirror
(source)
Current status and future plans
Our lab has made significant strides in protocol optimization for probe design, probe cleavage, 3’ end RNA library preparation, and raw data processing. Moving forward, we're focused on refining tissue processing protocols and developing comprehensive bioinformatics pipelines for seamless data analysis. Stay tuned!