Development of a digital PCR TERT/TERRA quantification assay in neuroblastoma
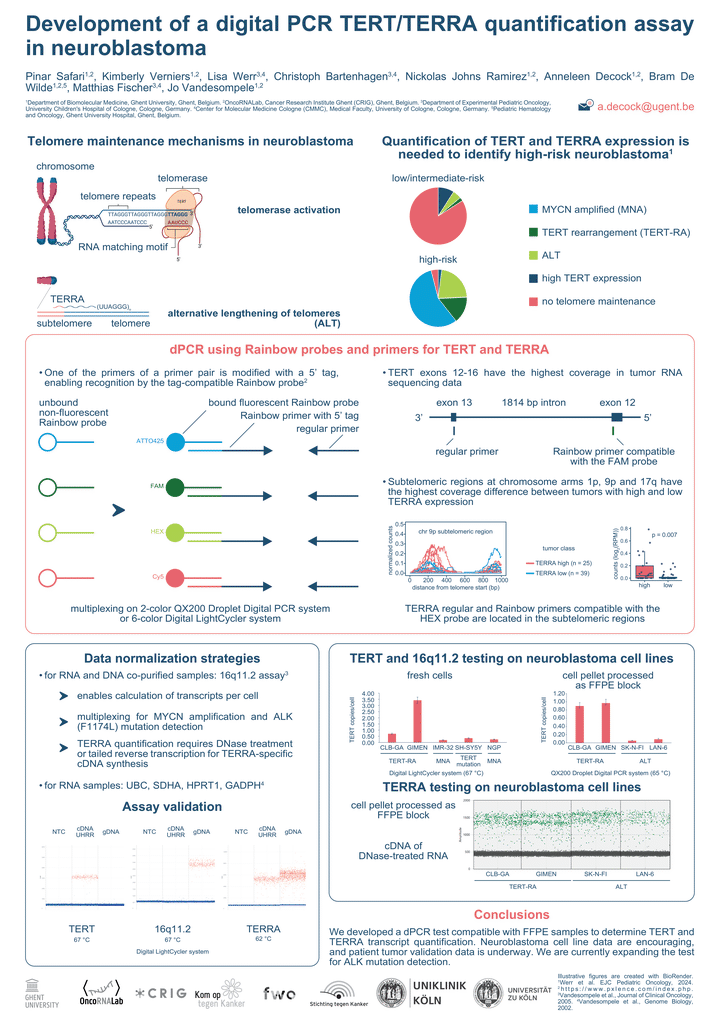
BACKGROUND: Telomere maintenance mechanisms are a hallmark of high-risk neuroblastoma, often characterized by overexpression of telomerase reverse transcriptase (TERT) and non-coding telomeric repeat–containing RNA (TERRA) transcripts.
AIMS: The development of a digital PCR assay for sensitive quantification of TERT and TERRA transcript copies per cell to allow risk stratification in neuroblastoma.
METHODS: Relevant TERRA loci (1p, 9p and 17q) were determined by having a higher RNA-seq coverage at subtelomeric regions in TERRA high vs. low neuroblastomas. Based on copy number profiles, a region on 16q11.2 was selected as having the least frequent copy number alterations, suitable for normalization of gene expression data and transcript counts per cell. Digital PCR assays were designed using Primer3Plus, verified for specificity and absence of secondary structures and ordered as Rainbow primers. DNA and RNA were co-purified from fresh-frozen and FFPE specimens before digital PCR was performed.
RESULTS: Assays where optimized by annealing temperature gradient analysis on human reference DNA and RNA and validated by preparing FFPE samples from neuroblastoma cells. We included CLBGA and GIMEN (TERT rearrangement =TERT-positive) and SKNFI and LAN6 (ALT-positive =TERT-negative). Our digital PCR test indicates that CLBGA and GIMEN have ~1.5-2 TERT RNA copies per cell, 30 times more than SKNFI and LAN6 cells (1 TERT transcript per ~27 and ~12 cells), suggesting that the assay can discriminate between TERT-positive and -negative samples. The test is currently converted into a multiplex test (also including ALK mutation and MYCN copy number), and tested on a large series of neuroblastomas with known TERT and TERRA status.
CONCLUSIONS: We developed a digital PCR test compatible with archived tissue samples to determine absolute TERT and TERRA transcripts counts per cell. Preliminary data are encouraging, and extended validation data is underway. In principle, the test can be easily expanded by including other targets or used for different tumor types.