Cancer type and patient-specific changes in pan-cancer blood plasma cfRNA profiles
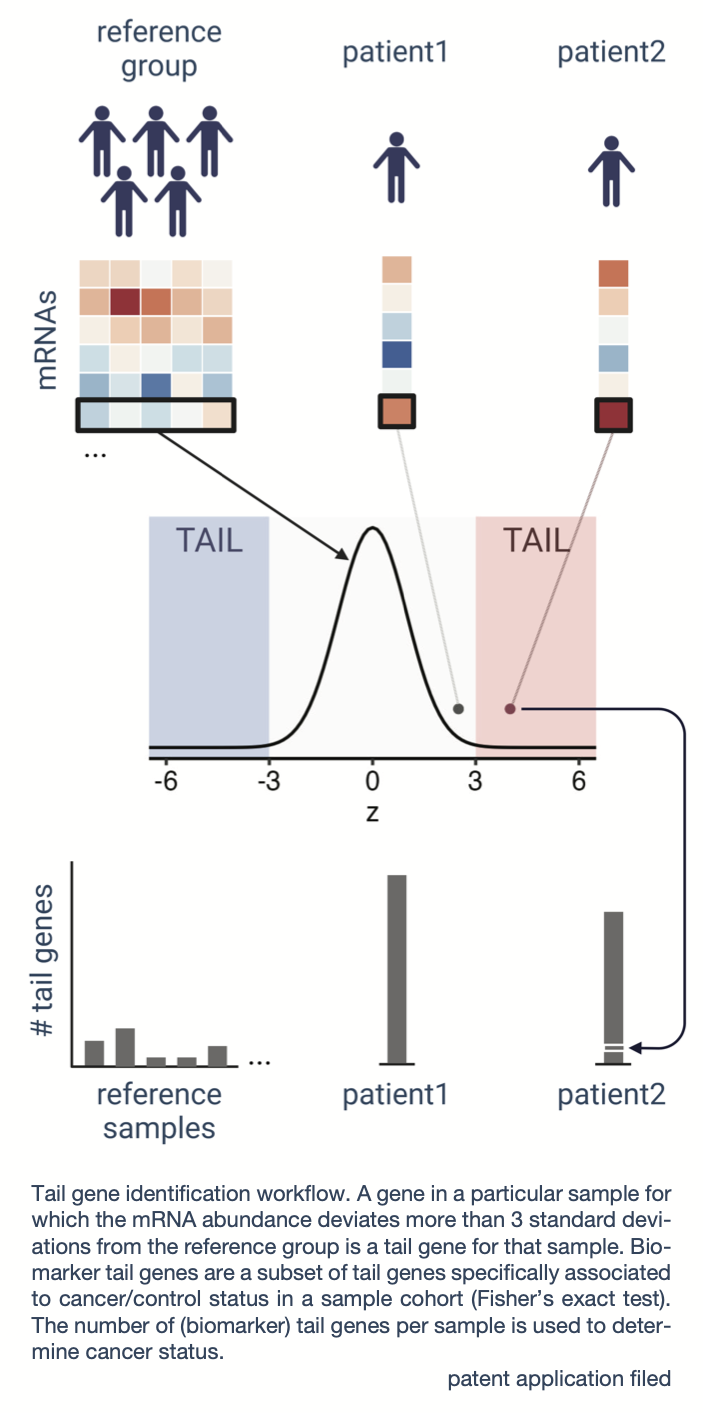
Circulating nucleic acids in blood plasma form an attractive resource to study human health and disease. While the presence of extracellular RNAs in plasma has been firmly established, their origin and clinical utility for management of cancer patients is less understood. Here, we applied mRNA capture sequencing of blood plasma cell-free RNA from 236 cancer patients representing 25 different locally advanced to metastatic cancer types and 30 cancer-free controls. We observed cancer-type specific as well as pan-cancer alterations in cell-free transcriptomes compared to controls, including tumor-derived RNAs, RNAs reflecting the tumor tissue-of-origin and RNAs from various immune cell types. However, differentially abundant RNAs were very heterogenous between patients and between cohorts, hampering identification of robust cancer biomarkers. To this end, we developed a novel method that compares each individual cancer patient to a reference control population to identify what we call tail biomarker genes. We demonstrate that these tail biomarker genes can discriminate patients from controls with very high accuracy. Our results were confirmed in an independent cohort of 65 plasma samples and in 24 urine samples. Together, our findings demonstrate heterogeneity in cell-free RNA alterations between cancer patients and indicate that case-specific alterations can be exploited for classification purposes.