A 3’-end capture sequencing method for high-throughput targeted gene expression profiling
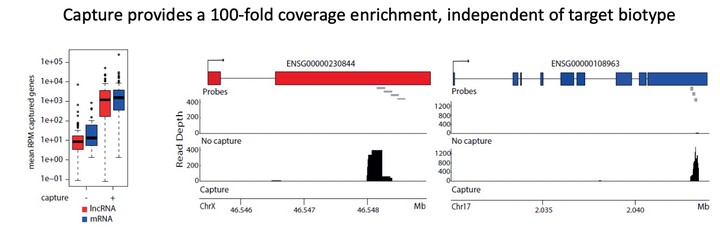
A 3’-end capture sequencing method for high-throughput targeted gene expression profiling Molecular phenotyping through shallow 3’-end RNA-sequencing workflows is increasingly applied in the context of large-scale chemical or genetic perturbation screens to study disease biology or support drug discovery. While these workflows enable accurate quantification of the most abundant genes, they are less effective for applications that require expression profiling of low abundant transcripts, like long non-coding RNAs (lncRNAs), or selected gene panels. To tackle these issues, we describe a workflow combining 3’-end library preparation with 3’-end hybrid capture probes and shallow RNA-sequencing for cost-effective, targeted quantification of subsets of (low abundant) genes across hundreds to thousands of samples. To assess the performance of the method, we designed a capture probe set for more than 100 mRNA and lncRNA target genes and applied the workflow to a cohort of 360 samples (lysates of HEK293T cells). When compared to standard 3’-end RNA-sequencing, 3’-end capture sequencing resulted in a more than 200-fold enrichment of target gene abundance while conserving relative inter-gene and inter-sample abundances and this with a very straightforward protocol. 80% of the total amount of reads are allocated to captured genes, making this a very effective technique. 3’-end RNA capture sequencing enables accurate targeted gene expression profiling at extremely shallow sequencing depth, resulting in a drastically lower sequencing cost.