Whole transcriptome profiling of liquid biopsies from tumor xenografted mouse models enables specific monitoring of tumor-derived extracellular RNA
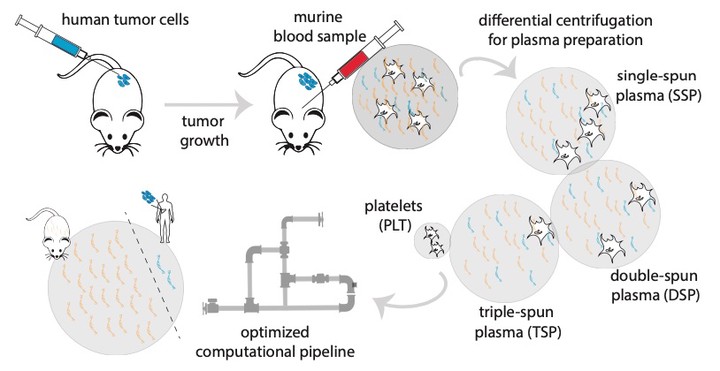
While cell-free DNA (cfDNA) is widely being investigated, free circulating RNA (extracellular RNA, exRNA) has the potential to improve cancer therapy response monitoring and detection due to its dynamic nature. However, it remains unclear in which blood subcompartment tumour-derived exRNAs primarily reside. We developed a host-xenograft deconvolution framework, exRNAxeno, with mapping strategies to either a combined human-mouse reference genome or both species genomes in parallel, that can be applied to exRNA sequencing data from liquid biopsies of human xenograft mouse models, enabling to distinguish (human) tumoural RNA from (murine) host RNA, and as such to specifically analyse tumour-derived exRNA. Subsequently, the combined pipeline was applied to total exRNA sequencing data from platelets and three plasma fractions from two xenograft models. Tumoural exRNA concentrations are not determined by plasma platelet levels, while host exRNA concentrations increase with platelet content. Furthermore, a large variability in exRNA levels and gene content across individual mice is observed. The tumoural gene detectability in plasma is largely correlated with the RNA expression levels in the tumour tissue or cell line. These findings unravel new aspects of tumour-derived exRNA biology in xenograft models and open new avenues to further investigate the role of exRNAs in cancer.