exRNAQC
 exRNAQC
exRNAQC
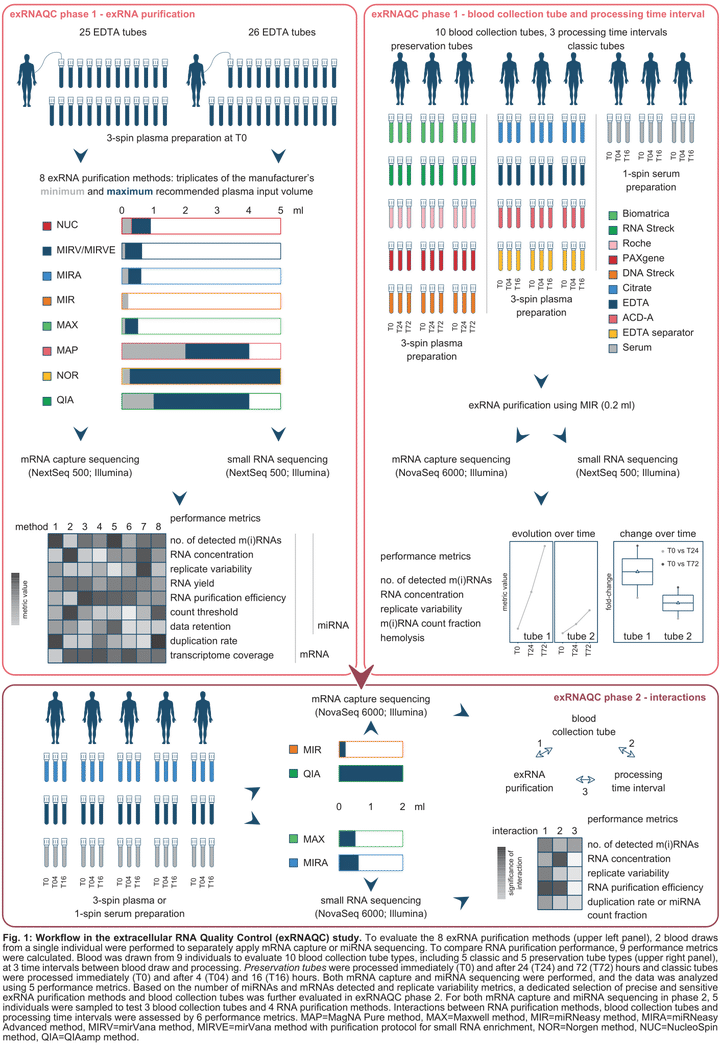
Blood-based extracellular RNA (cell-free RNA; exRNA) biomarkers require validated sample collection, processing, and quantification procedures. No study to date has systematically tested pre-analytical variables affecting transcriptome-wide exRNA analysis. By evaluating their impact on deep transcriptome profiling of microRNAs and mRNAs in blood plasma or serum, we compared ten blood collection tubes, three blood processing time intervals, and eight RNA purification methods. In addition, we assessed interactions among a selected pre-analytical variable set, resulting in 456 extracellular transcriptomes. Blood preservation tubes failed to stabilize exRNA and RNA purification methods differed significantly in performance, causing variations in concentration, detected gene numbers, replicability and observed transcriptome complexity. Critical interactions between tubes, purification methods and time intervals were identified. We provide 11 analytical performance metrics for exRNA quantification methods and put forward recommendations for both users and manufacturers of RNA purification methods and blood collection tubes, collectively, essential groundwork for exRNA-based precision medicine applications.
Detailed results are available here.
The exRNAQC Consortium
Author contributions are reported according to the CRediT taxonomy. Within the exRNAQC Consortium member list and the CRediT groups, authors are in alphabetical order: Anckaert J, Avila Cobos F, Decock A, Decruyenaere P, Deleu J, De Preter K, De Wever O, De Wilde J, Dhondt B, D’huyvetter T, Everaert C, Fierro C, Helsmoortel HH, Hendrix A, Hulstaert E, Koster J, Kuersten S, Mercer TR, Mestdagh P, Morlion A, Nijs N, Nuytens J, Philippron A, Piofczyk T, Poma-Soto F, Schoofs K, Schroth GP, Thas O, Vanden Eynde E, Vandesompele J, Van Maerken T, Van Paemel R, Verniers K, Verwilt J, Yigit N. Corresponding authors: Vandesompele J, Mestdagh P.