Digital PCR-based evaluation of nucleic acid extraction kit performance for the co-purification of cell-free DNA and RNA
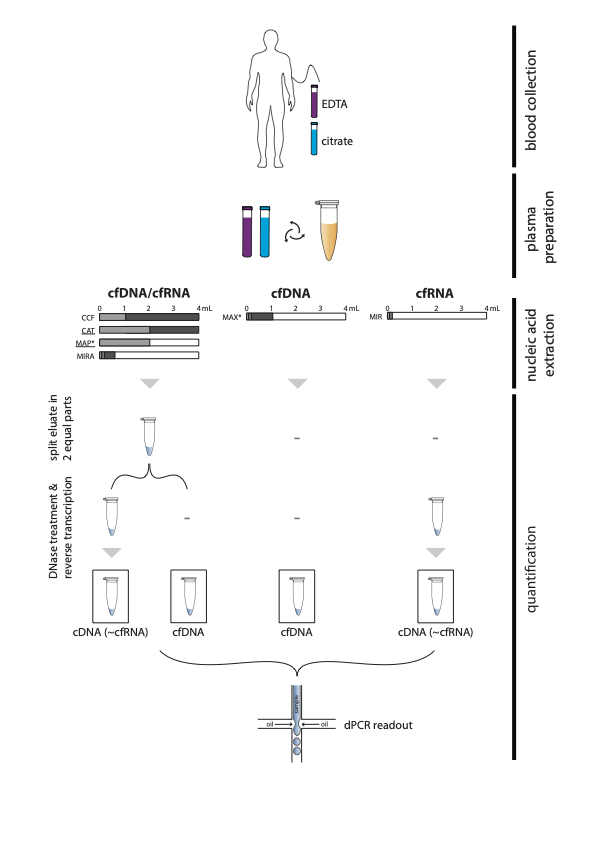 Experimental design to evaluate (co-)purification kits.
Experimental design to evaluate (co-)purification kits.
Abstract
Blood plasma, one of the most studied liquid biopsies, contains various molecules that have biomarker potential for cancer detection, including cell-free DNA (cfDNA) and cell-free RNA (cfRNA). As the vast majority of cell-free nucleic acids in circulation are non-cancerous, a laboratory workflow with a high detection sensitivity of tumor-derived nucleic acids is a prerequisite for precision oncology. One way to meet this requirement is by the combined analysis of cfDNA and cfRNA from the same liquid biopsy sample. So far, no study has systematically compared the performance of cfDNA and cfRNA co-purification to increase sensitivity. First, we set up a framework using digital PCR (dPCR) technology to quantify cfDNA and cfRNA from human blood plasma in order to compare cfDNA/cfRNA co-purification kit performance. To that end, we optimized two dPCR duplex assays, designed to quantify both cfDNA and cfRNA with the same assays, by ensuring that primers and probes are located within a highly abundant exon. Next, we applied our optimized workflow to evaluate the co-purification performance of two manual and two semi-automated methods over a range of plasma input volumes (0.06–4mL). Some kits result in higher nucleic acid concentrations in the eluate, while consuming only half of the plasma volume. The combined nucleic acid quantification systematically results in higher nucleic acid concentrations as compared to a parallel quantification of cfDNA and cfRNA in the eluate. We provide a framework to evaluate the performance of cfDNA/cfRNA co-purification kits and have tested two manual and two semi-automated co-purification kits in function of the available plasma input amount and the intended use of the nucleic acid eluate. We demonstrate that the combined quantification of cfDNA and cfRNA has a benefit compared to separate quantification. We foresee that the results of this study are instrumental for clinical applications to help increase mutation detection sensitivity, allowing improved disease detection and monitoring.