The neuroblastoma specific lincRNA NESPR controls noradrenergic cell identity
Abstract
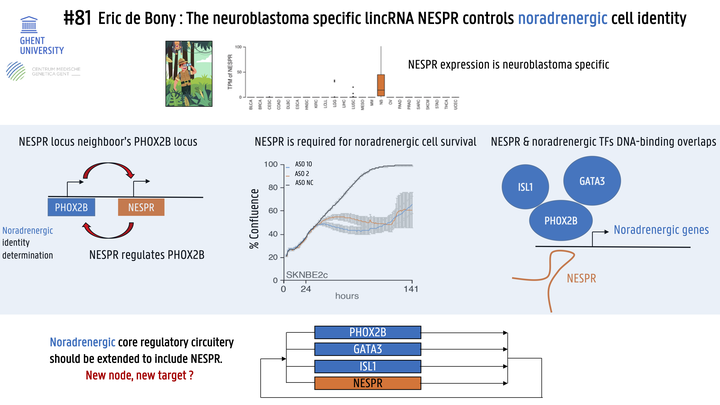
Neuroblastoma tumor cells are defined by a noradrenergic or mesenchymal state. Transcription factors composing the core regulatory circuitries (CRCs) that define these states are well described and include PHOX2B, ISL1, GATA3 and HAND2 for the noradrenergic state and FOSL1, FOSL2 and JUN for the mesenchymal state. The role of long non-coding RNAs (lncRNAs) in these CRCs remains poorly understood despite the fact that these molecules are emerging as transcription regulators. By studying the role that lncRNAs occupy in determining neuroblastoma cell identity and their relationship with protein-coding transcription factors we hope to paint a more accurate and comprehensive picture of core regulatory networks. We performed a pan-cancer lncRNA expression analysis comparing over 35 different tumor types to identify neuroblastoma-specific lncRNAs. Antisense oligonucleotides (ASOs) and siRNAs were used to silence the identified neuroblastoma-specific transcript. Treated cells were then subjected to colony formation, cell growth and apoptosis assays followed by RNA-sequencing. We then used 4C-sequencing to chart potential interactions between relevant loci. Finally, Chromatin Immunoprecipitation by RNA-Pulldown (ChIRP) sequencing was performed to identify DNA binding regions of the identified lncRNA. Pan-cancer expression analysis identified the neuroblastoma-specific lncRNA NESPR (NEuroblastoma Specific Phox2B Regulatory RNA). NESPR is located in the PHOX2B super-enhancer region and, unlike many lncRNAs, NESPR is abundantly expressed, efficiently spliced, highly conserved in mammals and present in both the nucleus and the cytoplasm as observed with fractionation and single-cell expression data. In neuroblastoma, high NESPR expression is associated with high-stage disease, MYCN amplification and poor patient survival. Interestingly, NESPR expression is confined to noradrenergic neuroblastoma cells and downregulated upon de-differentiation to the mesenchymal state. Knockdown of NESPR significantly decreased colony formation and cell growth and lead to an increase in apoptosis. Of note, these effects were strictly dependent on the nuclear fraction of NESPR. RNA-sequencing of NESPR-silenced neuroblastoma cell lines revealed a significant reduction in the expression of PHOX2B and other components of the noradrenergic CRC, suggesting that NESPR may be involved in regulating PHOX2B expression and, as such, defining noradrenergic cell identity. 4C-sequencing demonstrated a long-range interaction between the NESPR locus and the PHOX2B promoter in noradrenergic cell lines but not in mesenchymal cell lines. Moreover, ChIRP-sequencing revealed binding of NESPR to DNA elements confined within the PHOX2B insulated neighborhood, suggesting NESPR regulates PHOX2B expression in cis. Strikingly, ChIRP-sequencing revealed binding of NESPR to various regions outside the PHOX2B locus. Motif analysis of these NESPR binding sites revealed a significant enrichment of GATA3, ISL1 and HAND2 binding motifs, suggesting a potential trans function for NESPR in the noradrenergic CRC. Experiments are ongoing to further validate these findings. Overall, our results have uncovered a key regulatory function for the lncRNA NESPR in the noradrenergic CRC and demonstrate its role in neuroblastoma cell survival. To further elucidate the molecular mechanism underlying NESPR function, we are identifying its protein binding partners and potential influence on the chromatin environment of its target genes. Finally, we have established a NESPR knock-out mouse model to further study NESPR function in vivo.