Large-scale benchmarking of circular RNA detection tools
Abstract
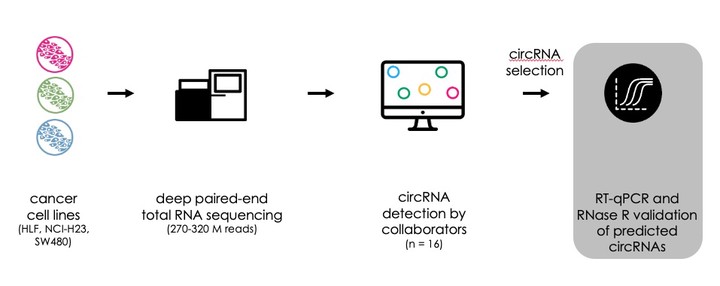
Over the last decade, many computational pipelines have been developed to identify circular RNA (circRNA) back-splice junctions (BSJ) in massively parallel RNA sequencing data. These circRNA detection pipelines widely differ with respect to their general approach (segmented-read versus candidate-based approach), alignment tool, and filtering steps. This leads to diverse circRNA predictions when using the same RNA sequencing data as input. To date, a gold standard for circRNA detection and quantification of RNA sequencing data is missing. To tackle this issue, we set up a collaborative benchmarking study involving the developers of 15 circRNA detection tools to identify the most accurate circRNA detection strategy. In a first phase, deep RNA sequencing data from RNase R treated and untreated human cancer cell line samples were shared with all collaborating tool developers. Comparing their circRNA prediction results in silico, substantial differences in the number of detected circRNAs were observed, ranging from less than 100 to more than 25,000 predicted circRNAs per sample. Some tools yield similar overall predictions, while others predict vastly different sets of circRNAs. In a second phase, the accuracy of the predictions of individual tools was assessed. For this, RT-qPCR of predicted BSJ on matched RNase R treated and untreated RNA was used as a gold standard. The exact sequence surrounding the BSJ position was confirmed by amplicon sequencing. In total, 1500 circRNAs were randomly selected from the extensive circRNA dataset (corresponding to ~100 circRNAs per tool) and used to determine the performance of each prediction tool. At the RNA society 2022 meeting, we will present data comparing each circRNA detection tool based on the estimated false-discovery rate and accuracy. In addition, we will provide guidelines for accurate circRNA detection in RNA sequencing data.