circRNA
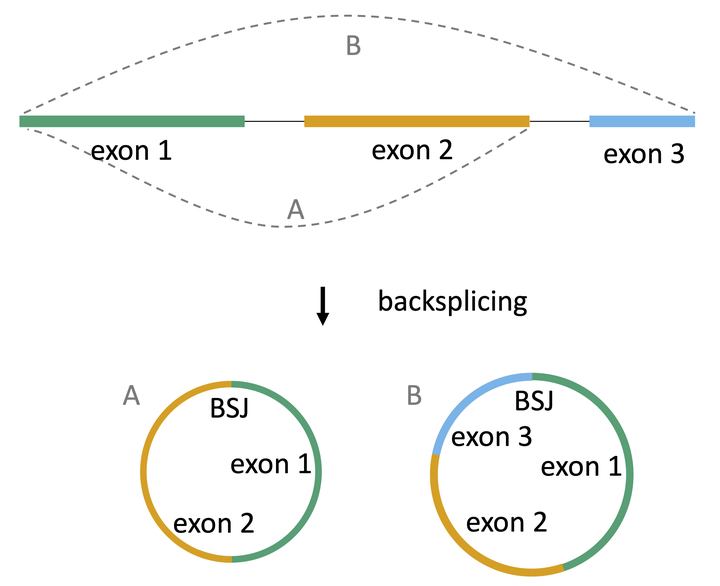 circRNA biogenesis
circRNA biogenesis
Introduction
After their discovery more than three decades ago, circular RNAs (circRNAs) have been emerging as a large class of generally non-coding RNAs. Originating from the same precursor as linear RNA transcripts, circRNAs are formed through a process called back-splicing, which results in a back-splice junction (BSJ) between a splice donor and an upstream splice acceptor. Due to their circular nature, circRNAs are more resistant to degradation by exonucleases and therefore more stable than linear RNA 1 2. CircRNAs are widespread and abundant in a variety of organisms. Interestingly, the majority of circRNAs seem to be cell-type specific 3 4.
CircRNA detection
CircRNAs are typically identified in RNA sequencing data (RNA-seq) and numerous computational circRNA identification pipelines data have been developed 5678. These pipelines differ in their choice of alignment tool, the use of gene annotation, the ability use of single-end or paired-end data, and in their filtering steps. Recently, we published a circRNA detection tool benchmarking in which 16 different tools were compared and validated. The benchmarking was published in Nature Methods and was also discussed in a Research Briefing and a GenomeWeb news article.
However, these circRNA detection tools are based on short-read sequencing techniques and lack the ability to accurately detect full-length circRNA sequences. In our lab, Jasper Verwilt is looking into long read sequencing methods and their possible application for the detection of circRNAs. As the internal sequence of most circRNAs remains unknown, there is a high demand for such techniques.
An alternative technique to detect circRNAs, is RT-qPCR with divergent primers. For these experiments, Steve Lefever successfully adapted his primer design tool PimerXL9 to design specific circRNA primers.
CircRNA databases
In addition to the circRNA prediction tools, various databases cataloguing circRNAs have been developed as well. In January 2020, Marieke Vromman and colleagues published a review presenting a comprehensive overview of the current circRNA databases and their content, features and usability. Furthermore, in this review they discuss the current issues regarding circRNA databases and come with important suggestions to streamline further research in this growing field 10.
CircRNAs in cancer
Interestingly, circRNAs have been associated with a broad range of diseases, including various types of cancer 11 12. Due to the observed associations between circRNA abundance and cancer, circRNAs may serve as cancer biomarkers with good diagnostic performance 13. Various studies demonstrated that circRNAs are present at relatively high steady state levels in human biofluids, such as saliva, plasma, serum and in exosomes, which makes them attractive candidate biomarkers for non-invasive liquid biopsies 1. In our lab, Eva Hulstaert is looking into the circRNA content in 20 different human biofluids using mRNA capture sequencing 14. Furthermore, Annelien Morlion and Kathleen Schoofs are studying circRNAs in plasma from patients with cancer (26 different cancer types) and plasma from healthy donors.
On the other hand, Lucía Lorenzi set up the RNA Atlas 15, a single nucleotide resolution map of the human transcriptome consisting of matching small, polyA and total RNA seq profiles of a heterogeneous collection of nearly 300 different human tissues and cell types. Using this extensive total RNA seq dataset, circRNA detection was performed and 37,140 circRNAs were discovered in human cell and tissues.
CircRNA primer design
In general, large-scale circRNA BSJ detection is performed based on RNA sequencing data, followed by the selection and validation of circRNAs of interest using RT-qPCR with circRNA-specific PCR primers. Such a primer pair is convergent and functional on the circRNA template but divergent and non-functional on the linear host gene. Although a few circRNA primer design pipelines have been published, none of them offer large-scale, easy-to-use circRNA primer design. Other limitations are that these tools generally do not take into account assay specificity, secondary structures, and SNPs in the primer annealing regions. Furthermore, these tools are limited to circRNA primer design for humans (no other organisms possible), and no wet-lab validation is demonstrated. Here, we present CIRCprimerXL, a circRNA RT-qPCR assay design pipeline based on the primer design framework primerXL. CIRCprimerXL takes a circRNA BSJ position as input, and designs BSJ-spanning primers using Primer3. The user can choose to use the unspliced or spliced circRNA sequence as template. Prior to primer design, sequence regions with secondary structures and common SNPs are flagged. Next, the primers are filtered based on predicted specificity and the absence of secondary structures of the amplicon to select a suitable primer pair. Our tool is both available as a user-friendly web tool and as a stand-alone pipeline based on Docker and Nextflow, allowing users to run the pipeline on a wide range of computer infrastructures. The CIRCprimerXL Nextflow pipeline can be used to design circRNA primers for any species by providing the appropriate reference genome. The CIRCprimerXL web tool supports circRNA primer design for human, mouse, rat, zebrafish, Xenopus tropicalis, and C. elegans. The design process can easily be scaled up for the qPCR assay design of tens of thousands of circRNAs within a couple of hours. We show how CIRCprimerXL has been successfully used to design qPCR assays for over 15,000 human circRNAs of which 20 were empirically validated. CIRCprimerXL software, documentation, and test data can be found at: https://github.com/OncoRNALab/CIRCprimerXL. CIRCprimerXL is also implemented as a webtool at: https://circprimerxl.cmgg.be.
References
-
Su, M. et al. Circular RNAs in Cancer: Emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Molecular Cancer vol. 18 90 (2019). ↩︎ ↩︎
-
Salzman, J., Gawad, C., Wang, P. L., Lacayo, N. & Brown, P. O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 7, e30733 (2012). ↩︎
-
Salzman, J., Chen, R. E., Olsen, M. N., Wang, P. L. & Brown, P. O. Cell-Type Specific Features of Circular RNA Expression. PLoS Genet. 9, e1003777 (2013). ↩︎
-
Hang, R. et al. Comprehensive characterization of circular RNAs in ~1000 human cancer cell lines. Genome Med. 11, 55 (2019). ↩︎
-
Gao, Y. & Zhao, F. Computational Strategies for Exploring Circular RNAs. Trends Genet. 34, 389–400 (2018). ↩︎
-
Hansen, T. B., Venø, M. T., Damgaard, C. K. & Kjems, J. Comparison of circular RNA prediction tools. Nucleic Acids Res. 44, e58 (2015). ↩︎
-
Zeng, X., Lin, W., Guo, M. & Zou, Q. A comprehensive overview and evaluation of circular RNA detection tools. PLoS Comput. Biol. 13, e1005420 (2017). ↩︎
-
Jakobi, T. & Dieterich, C. Computational approaches for circular RNA analysis. Wiley Interdiscip. Rev. RNA 2019, e1528 (2019). ↩︎
-
Lefever, S. et al. High-throughput PCR assay design for targeted resequencing using primerXL. BMC Bioinformatics 18, 1–9 (2017). ↩︎
-
Vromman, M., Vandesompele, J. & Volders, P.-J. Closing the circle: current state and perspectives of circular RNA databases. Brief. Bioinform. (2020) doi:10.1093/bib/bbz175. ↩︎
-
Vo, J. N. et al. The Landscape of Circular RNA in Cancer. Cell 176, 869–881 (2019). ↩︎
-
Shang, Q., Yang, Z., Jia, R. & Ge, S. The novel roles of circRNAs in human cancer. Mol. Cancer 18, 6 (2019). ↩︎
-
Tan, H., Gan, L., Fan, X., Liu, L. & Liu, S. Diagnostic value of circular RNAs as effective biomarkers for cancer: A systematic review and meta-analysis. Onco. Targets. Ther. 12, 2623–2633 (2019). ↩︎
-
Hulstaert, E. et al. Charting extracellular transcriptomes in The Human Biofluid RNA Atlas. bioRxiv (2019) doi:10.1017/CBO9781107415324.004. ↩︎
-
Lorenzi, L. et al. The RNA Atlas, a single nucleotide resolution map of the human transcriptome. bioRxiv 807529 (2019) doi:10.1101/807529. ↩︎