mRNA capture sequencing and RT-qPCR for the detection of pathognomonic, novel, and secondary fusion transcripts in FFPE tissue: a sarcoma showcase
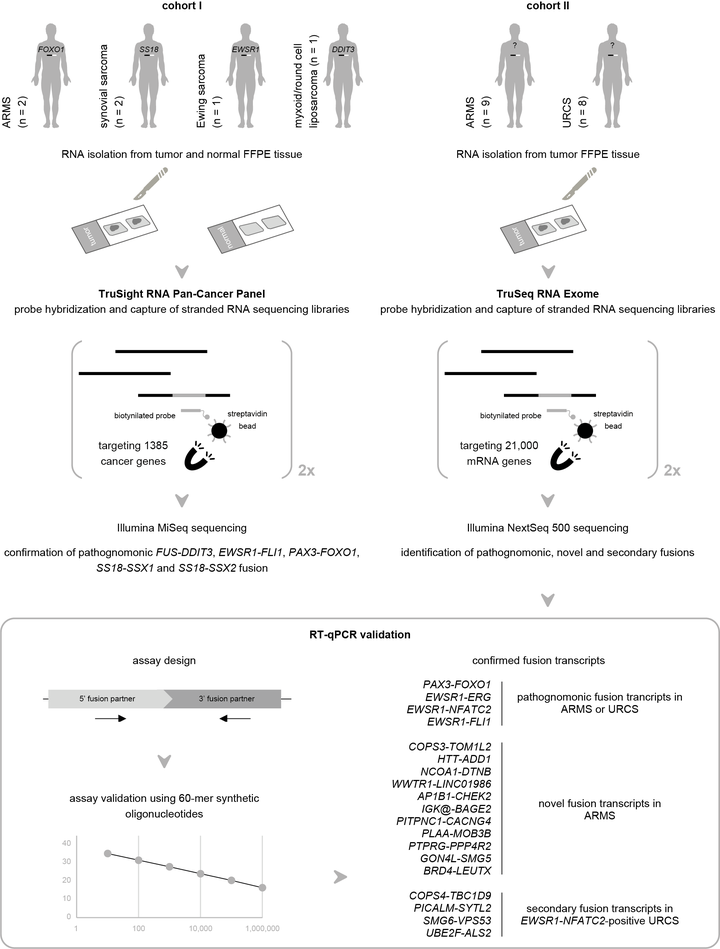 mRNA capture sequencing and RT-qPCR for the detection of pathognomonic, novel, and secondary fusion transcripts in FFPE sarcoma tissue. Cohort I, comprising six patients with a known pathognomonic fusion, is profiled using the TruSight RNA Pan-Cancer Panel. These data validated the mRNA capture sequencing analysis workflow for the identification of fusion transcripts. Subsequently, a second cohort of sarcomas that were designated fusion gene-negative by FISH analysis was analyzed using TruSeq RNA Exome sequencing. Multiple pathognomonic, novel, and secondary fusion transcripts were picked up and confirmed by RT-qPCR.
mRNA capture sequencing and RT-qPCR for the detection of pathognomonic, novel, and secondary fusion transcripts in FFPE sarcoma tissue. Cohort I, comprising six patients with a known pathognomonic fusion, is profiled using the TruSight RNA Pan-Cancer Panel. These data validated the mRNA capture sequencing analysis workflow for the identification of fusion transcripts. Subsequently, a second cohort of sarcomas that were designated fusion gene-negative by FISH analysis was analyzed using TruSeq RNA Exome sequencing. Multiple pathognomonic, novel, and secondary fusion transcripts were picked up and confirmed by RT-qPCR.
Abstract
We assess the performance of mRNA capture sequencing to identify fusion transcripts in FFPE tissue of different sarcoma types, followed by RT-qPCR confirmation. To validate our workflow, six positive control tumors with a specific chromosomal rearrangement were analyzed using the TruSight RNA Pan-Cancer Panel. Fusion transcript calling by FusionCatcher confirmed these aberrations and enabled the identification of both fusion gene partners and breakpoints. Next, whole-transcriptome TruSeq RNA Exome sequencing was applied to 17 fusion gene-negative alveolar rhabdomyosarcoma (ARMS) or undifferentiated round cell sarcoma (URCS) tumors, for whom fluorescence in situ hybridization (FISH) did not identify the classical pathognomonic rearrangements. For six patients, a pathognomonic fusion transcript was readily detected, i.e., PAX3-FOXO1 in two ARMS patients, and EWSR1-FLI1, EWSR1-ERG, or EWSR1-NFATC2 in four URCS patients. For the 11 remaining patients, 11 newly identified fusion transcripts were confirmed by RT-qPCR, including COPS3-TOM1L2, NCOA1-DTNB, WWTR1-LINC01986, PLAA-MOB3B, AP1B1-CHEK2, and BRD4-LEUTX fusion transcripts in ARMS patients. Additionally, recurrently detected secondary fusion transcripts in patients diagnosed with EWSR1-NFATC2-positive sarcoma were confirmed (COPS4-TBC1D9, PICALM-SYTL2, SMG6-VPS53, and UBE2F-ALS2). In conclusion, this study shows that mRNA capture sequencing enhances the detection rate of pathognomonic fusions and enables the identification of novel and secondary fusion transcripts in sarcomas.